$$
\newcommand{\mybold}[1]{\boldsymbol{#1}}
\newcommand{\trans}{\intercal}
\newcommand{\norm}[1]{\left\Vert#1\right\Vert}
\newcommand{\abs}[1]{\left|#1\right|}
\newcommand{\bbr}{\mathbb{R}}
\newcommand{\bbz}{\mathbb{Z}}
\newcommand{\bbc}{\mathbb{C}}
\newcommand{\gauss}[1]{\mathcal{N}\left(#1\right)}
\newcommand{\chisq}[1]{\mathcal{\chi}^2_{#1}}
\newcommand{\studentt}[1]{\mathrm{StudentT}_{#1}}
\newcommand{\fdist}[2]{\mathrm{FDist}_{#1,#2}}
\newcommand{\argmin}[1]{\underset{#1}{\mathrm{argmin}}\,}
\newcommand{\projop}[1]{\underset{#1}{\mathrm{Proj}}\,}
\newcommand{\proj}[1]{\underset{#1}{\mybold{P}}}
\newcommand{\expect}[1]{\mathbb{E}\left[#1\right]}
\newcommand{\prob}[1]{\mathbb{P}\left(#1\right)}
\newcommand{\dens}[1]{\mathit{p}\left(#1\right)}
\newcommand{\var}[1]{\mathrm{Var}\left(#1\right)}
\newcommand{\cov}[1]{\mathrm{Cov}\left(#1\right)}
\newcommand{\sumn}{\sum_{n=1}^N}
\newcommand{\meann}{\frac{1}{N} \sumn}
\newcommand{\cltn}{\frac{1}{\sqrt{N}} \sumn}
\newcommand{\trace}[1]{\mathrm{trace}\left(#1\right)}
\newcommand{\diag}[1]{\mathrm{Diag}\left(#1\right)}
\newcommand{\grad}[2]{\nabla_{#1} \left. #2 \right.}
\newcommand{\gradat}[3]{\nabla_{#1} \left. #2 \right|_{#3}}
\newcommand{\fracat}[3]{\left. \frac{#1}{#2} \right|_{#3}}
\newcommand{\W}{\mybold{W}}
\newcommand{\w}{w}
\newcommand{\wbar}{\bar{w}}
\newcommand{\wv}{\mybold{w}}
\newcommand{\X}{\mybold{X}}
\newcommand{\x}{x}
\newcommand{\xbar}{\bar{x}}
\newcommand{\xv}{\mybold{x}}
\newcommand{\Xcov}{\Sigmam_{\X}}
\newcommand{\Xcovhat}{\hat{\Sigmam}_{\X}}
\newcommand{\Covsand}{\Sigmam_{\mathrm{sand}}}
\newcommand{\Covsandhat}{\hat{\Sigmam}_{\mathrm{sand}}}
\newcommand{\Z}{\mybold{Z}}
\newcommand{\z}{z}
\newcommand{\zv}{\mybold{z}}
\newcommand{\zbar}{\bar{z}}
\newcommand{\Y}{\mybold{Y}}
\newcommand{\Yhat}{\hat{\Y}}
\newcommand{\y}{y}
\newcommand{\yv}{\mybold{y}}
\newcommand{\yhat}{\hat{\y}}
\newcommand{\ybar}{\bar{y}}
\newcommand{\res}{\varepsilon}
\newcommand{\resv}{\mybold{\res}}
\newcommand{\resvhat}{\hat{\mybold{\res}}}
\newcommand{\reshat}{\hat{\res}}
\newcommand{\betav}{\mybold{\beta}}
\newcommand{\betavhat}{\hat{\betav}}
\newcommand{\betahat}{\hat{\beta}}
\newcommand{\betastar}{{\beta^{*}}}
\newcommand{\bv}{\mybold{\b}}
\newcommand{\bvhat}{\hat{\bv}}
\newcommand{\alphav}{\mybold{\alpha}}
\newcommand{\alphavhat}{\hat{\av}}
\newcommand{\alphahat}{\hat{\alpha}}
\newcommand{\omegav}{\mybold{\omega}}
\newcommand{\gv}{\mybold{\gamma}}
\newcommand{\gvhat}{\hat{\gv}}
\newcommand{\ghat}{\hat{\gamma}}
\newcommand{\hv}{\mybold{\h}}
\newcommand{\hvhat}{\hat{\hv}}
\newcommand{\hhat}{\hat{\h}}
\newcommand{\gammav}{\mybold{\gamma}}
\newcommand{\gammavhat}{\hat{\gammav}}
\newcommand{\gammahat}{\hat{\gamma}}
\newcommand{\new}{\mathrm{new}}
\newcommand{\zerov}{\mybold{0}}
\newcommand{\onev}{\mybold{1}}
\newcommand{\id}{\mybold{I}}
\newcommand{\sigmahat}{\hat{\sigma}}
\newcommand{\etav}{\mybold{\eta}}
\newcommand{\muv}{\mybold{\mu}}
\newcommand{\Sigmam}{\mybold{\Sigma}}
\newcommand{\rdom}[1]{\mathbb{R}^{#1}}
\newcommand{\RV}[1]{\tilde{#1}}
\def\A{\mybold{A}}
\def\A{\mybold{A}}
\def\av{\mybold{a}}
\def\a{a}
\def\B{\mybold{B}}
\def\S{\mybold{S}}
\def\sv{\mybold{s}}
\def\s{s}
\def\R{\mybold{R}}
\def\rv{\mybold{r}}
\def\r{r}
\def\V{\mybold{V}}
\def\vv{\mybold{v}}
\def\v{v}
\def\U{\mybold{U}}
\def\uv{\mybold{u}}
\def\u{u}
\def\W{\mybold{W}}
\def\wv{\mybold{w}}
\def\w{w}
\def\tv{\mybold{t}}
\def\t{t}
\def\Sc{\mathcal{S}}
\def\ev{\mybold{e}}
\def\Lammat{\mybold{\Lambda}}
$$
\(\,\)
library (tidyverse)library (sandwich)theme_update (text = element_text (size= 24 ))options (repr.plot.width= 12 , repr.plot.height= 6 )
── Attaching core tidyverse packages ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.2 ✔ readr 2.1.4
✔ forcats 1.0.0 ✔ stringr 1.5.0
✔ ggplot2 3.4.2 ✔ tibble 3.2.1
✔ lubridate 1.9.2 ✔ tidyr 1.3.0
✔ purrr 1.0.1
── Conflicts ────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
<- FALSE # https://avehtari.github.io/ROS-Examples/examples.html if (install) {install.packages ("rprojroot" ):: install_github ("avehtari/ROS-Examples" , subdir= "rpackage" ) library (rosdata)data (kidiq)
A data.frame: 6 × 5
<int>
<int>
<dbl>
<int>
<int>
1
65
1
121.11753
4
27
2
98
1
89.36188
4
25
3
85
1
115.44316
4
27
4
83
1
99.44964
3
25
5
115
1
92.74571
4
27
6
98
0
107.90184
1
18
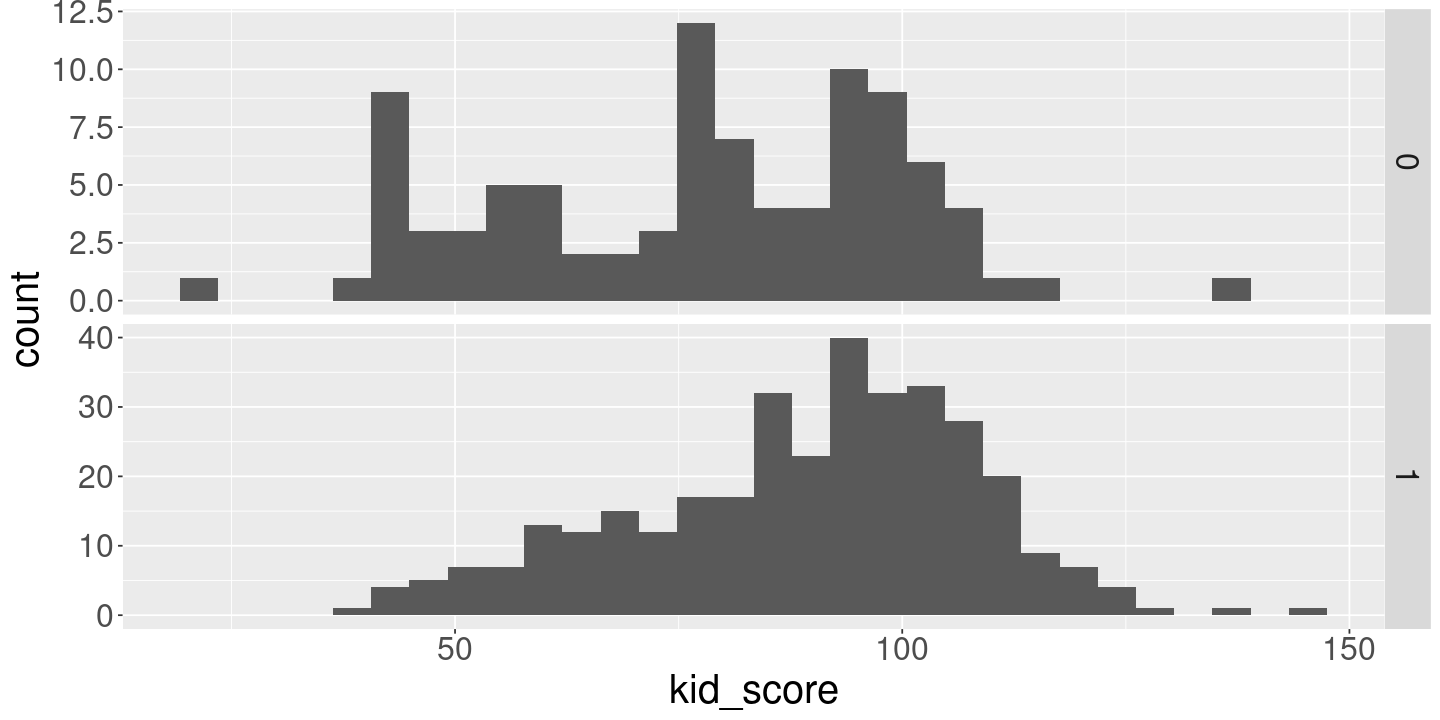
<- lm (kid_score ~ 1 + mom_hs, kidiq)print (fit_hs)ggplot (kidiq) + geom_histogram (aes (x= kid_score)) + facet_grid (mom_hs ~ ., scales= "free" )
Call:
lm(formula = kid_score ~ 1 + mom_hs, data = kidiq)
Coefficients:
(Intercept) mom_hs
77.55 11.77
`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
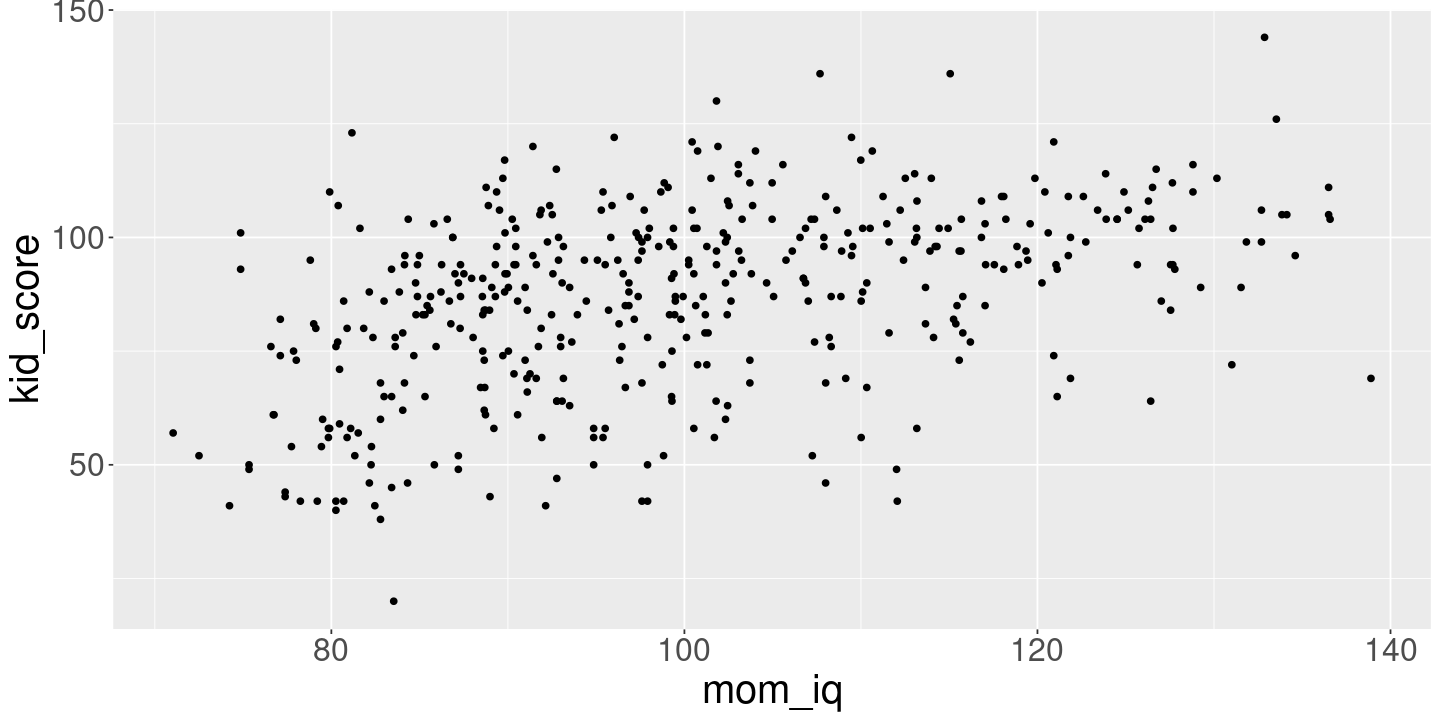
Kid score = \(\beta_0\) + \(\beta_1\) mom_iq
<- lm (kid_score ~ mom_iq, kidiq)print (fit_iq)ggplot (kidiq) + geom_point (aes (x= mom_iq, y= kid_score))
Call:
lm(formula = kid_score ~ mom_iq, data = kidiq)
Coefficients:
(Intercept) mom_iq
25.80 0.61
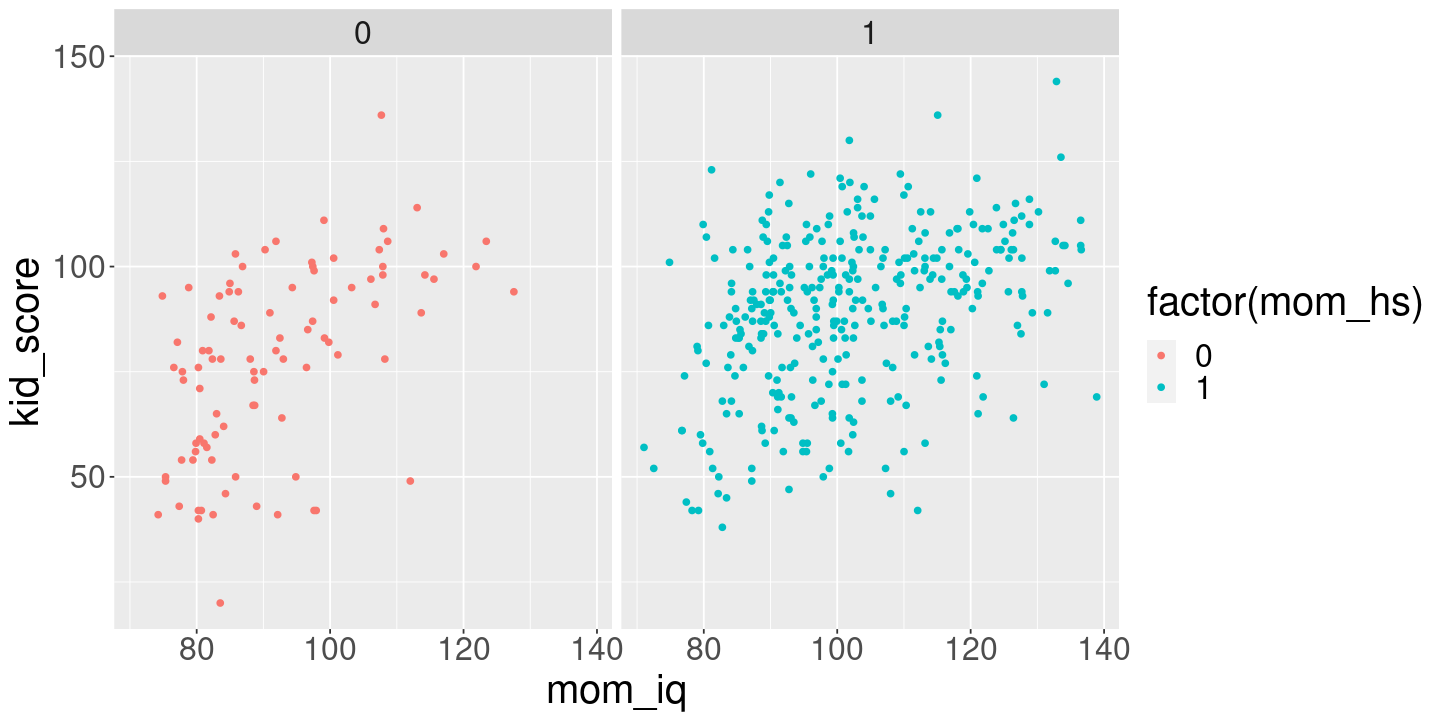
<- lm (kid_score ~ mom_hs + mom_iq, kidiq)ggplot (kidiq) + geom_point (aes (x= mom_iq, y= kid_score, color= factor (mom_hs), group= mom_hs)) + facet_grid (. ~ mom_hs)print (fit_hs_iq)
Call:
lm(formula = kid_score ~ mom_hs + mom_iq, data = kidiq)
Coefficients:
(Intercept) mom_hs mom_iq
25.7315 5.9501 0.5639
print (fit_hs)print (fit_iq)print (fit_hs_iq)
Call:
lm(formula = kid_score ~ 1 + mom_hs, data = kidiq)
Coefficients:
(Intercept) mom_hs
77.55 11.77
Call:
lm(formula = kid_score ~ mom_iq, data = kidiq)
Coefficients:
(Intercept) mom_iq
25.80 0.61
Call:
lm(formula = kid_score ~ mom_hs + mom_iq, data = kidiq)
Coefficients:
(Intercept) mom_hs mom_iq
25.7315 5.9501 0.5639
Simulations
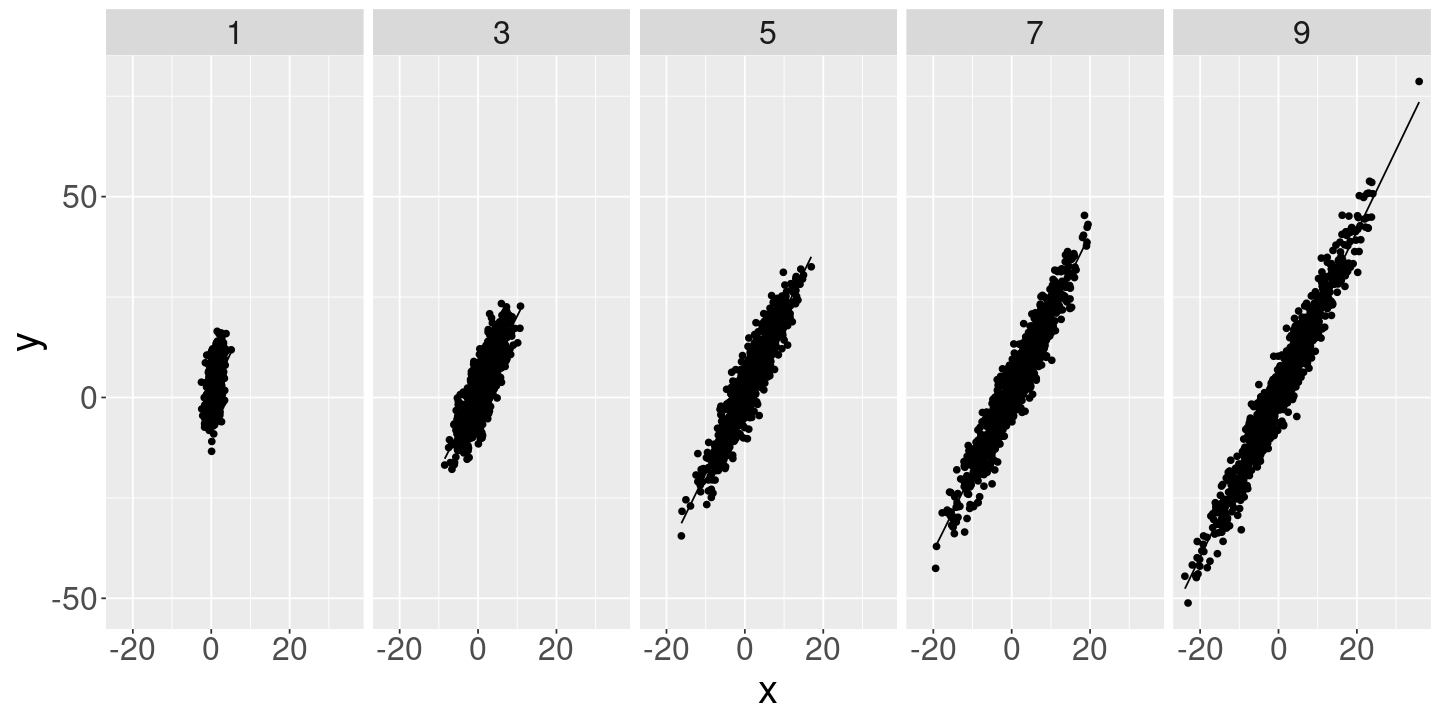
Let’s look at the behavior of the standard errors of \(\betahat_1\) when regressing \(\y_n \sim \beta_0 + \beta_1 \x_n\) , where \(\var{\res_n} = \sigma^2\) and \(\var{\x_n} = \sigma_x^2\) .
<- seq (1.0 , 10 , by= 2 )<- 4.0 <- 1000 <- 1.0 <- 2.0 <- list ()<- data.frame ()<- data.frame ()for (sigma_x in sigma_x_vals) {<- rnorm (n_obs, mean= 1.0 , sd= sigma_x)<- rnorm (n_obs, mean= 0 , sd= sigma_eps)<- beta_0 + beta_1 * x + eps<- data.frame (y= y, x= x, eps= eps, sigma_x= sigma_x)<- lm (y ~ 1 + x, df)<- summary (fit)$ coefficients["x" , , drop= FALSE ] %>% as.data.frame () %>% mutate (sigma_x= sigma_x)<- bind_rows (fits, coeffs)<- df %>% mutate (y_pred= fit$ fitted.values)<- bind_rows (df_comb, df)
ggplot (df_comb) + geom_point (aes (x= x, y= y)) + geom_line (aes (x= x, y= y_pred)) + facet_grid (~ sigma_x)
A data.frame: 2 × 5
<dbl>
<dbl>
<dbl>
<dbl>
<dbl>
x...1
1.983005
0.131168439
15.118
1.193266e-46
1
x...2
1.998115
0.004886705
408.888
0.000000e+00
26